hifiasm组装
# 介绍
Hifiasm 是一款针对PacBio Hifi reads单倍体解析的快速从头组装工具
仓库地址
https://github.com/chhylp123/hifiasm
文章
https://arxiv.org/pdf/2008.01237.pdf
# 软件安装及测试
# 安装方法
# Install hifiasm (requiring g++ and zlib)
git clone https://github.com/chhylp123/hifiasm
cd hifiasm && make
2
3
# 小测试数据下载
wget https://github.com/chhylp123/hifiasm/releases/download/v0.7/chr11-2M.fa.gz
大数据:小鼠数据
# 测试数据运行
针对小数据使用-f0
./hifiasm -o test -t4 -f0 chr11-2M.fa.gz 2> test.log
从gfa文件获得primary contigs
awk '/^S/{print ">"$2;print $3}' test.p_ctg.gfa > test.p_ctg.fa
# 软件参数
- -l0 :关掉 duplication purging,对于近交或者纯合基因组,建议加上该参数。
- -t :CPU 线程
- -o : 输出文件的前缀
- -z20 : 一些旧的HiFi reads可能会存在短的接头,使用该参数进行修剪
- -f0 : 对于小基因组,该参数可以关闭起始bloom filter(该步骤会在开始时消耗16G内存)
- -f38/-f39 : 如果基因组大于人类,建议使用该参数以节省内存
# Trio binning组装
Trio binning使用来自两个亲本基因组的reads,首先将来自后代的长读段划分为单倍型特异性集。然后将每个单体型独立组装,以完成完整的二倍体重建。
Trio binning assembly (requiring https://github.com/lh3/yak)
yak count -b37 -t16 -o pat.yak <(cat pat_1.fq.gz pat_2.fq.gz) <(cat pat_1.fq.gz pat_2.fq.gz)
yak count -b37 -t16 -o mat.yak <(cat mat_1.fq.gz mat_2.fq.gz) <(cat mat_1.fq.gz mat_2.fq.gz)
hifiasm -o HG002.asm -t32 -1 pat.yak -2 mat.yak HG002-HiFi.fa.gz
2
3
# 输出结果
# non-trio assembly
- Haplotype-resolved raw unitig graph in GFA format (prefix.r_utg.gfa). 保留了所有单倍体信息,包括一些体细胞突变和测序错误。
- Haplotype-resolved processed unitig graph without small bubbles (prefix.p_utg.gfa). 相对于raw unitig,去除了图上的小气泡;这些小气泡可能是由于体细胞突变和数据里噪音引起的,不是真正的单倍体信息
- Primary assembly contig graph (prefix.p_ctg.gfa). 折叠了不同的单倍型。
- Alternate assembly contig graph (prefix.a_ctg.gfa). This graph consists of all assemblies that are discarded in primary contig graph.
# trio assembly
- Haplotype-resolved raw unitig graph in GFA format (prefix.r_utg.gfa). This graph keeps all haplotype information.
- Phased paternal/haplotype1 contig graph (prefix.hap1.p_ctg.gfa). This graph keeps the phased paternal/haplotype1 assembly.
- Phased maternal/haplotype2 contig graph (prefix.hap2.p_ctg.gfa). This graph keeps the phased maternal/haplotype2 assembly.
Hifiasm writes error corrected reads to the prefix.ec.bin binary file and writes overlaps to prefix.ovlp.source.bin and prefix.ovlp.reverse.bin.
Purging haplotig duplications may introduce misassemblies.
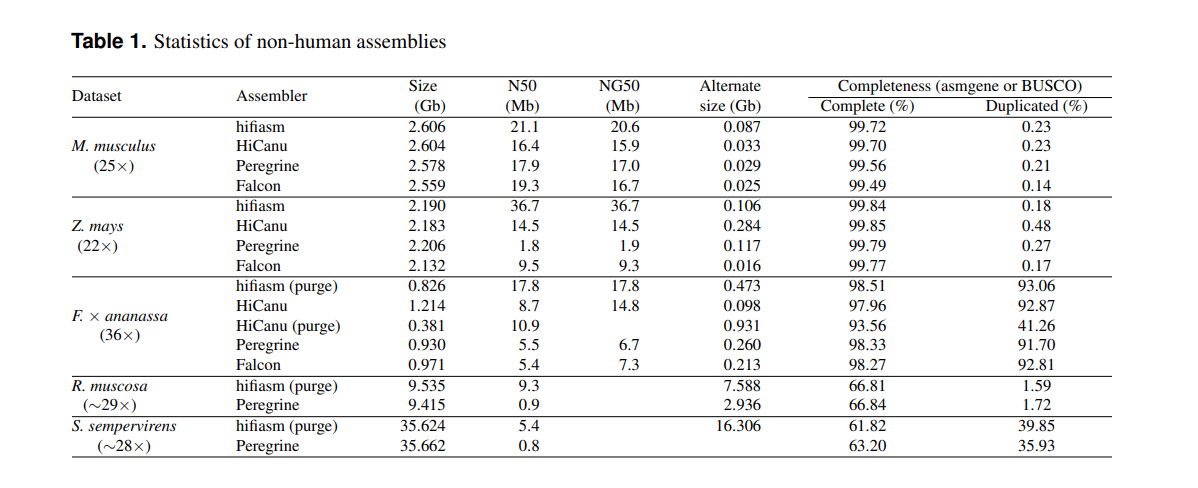
# 组装软件比较
# 其他
# HiFi 组装实例
The following table shows the statistics of several hifiasm primary assemblies:
| Dataset | Size | Cov. | Asm options | CPU time | Wall time | RAM | N50 |
|---|---|---|---|---|---|---|---|
| Mouse (C57/BL6J) | 2.6Gb | ×25 | -t48 -l0 | 172.9h | 4.8h | 76G | 21.1Mb |
| Maize (B73) | 2.2Gb | ×22 | -t48 -l0 | 203.2h | 5.1h | 68G | 36.7Mb |
| Strawberry | 0.8Gb | ×36 | -t48 -D10 | 152.7h | 3.7h | 91G | 17.8Mb |
| Frog | 9.5Gb | ×29 | -t48 | 2834.3h | 69.0h | 463G | 9.3Mb |
| Redwood | 35.6Gb | ×28 | -t80 | 3890.3h | 65.5h | 699G | 5.4Mb |
| Human (CHM13) | 3.1Gb | ×32 | -t48 -l0 | 310.7h | 8.2h | 114G | 88.9Mb |
| Human (HG00733) | 3.1Gb | ×33 | -t48 | 269.1h | 6.9h | 135G | 69.9Mb |
| Human (HG002) | 3.1Gb | ×36 | -t48 | 305.4h | 7.7h | 137G | 98.7Mb |
Hifiasm can assemble a 3.1Gb human genome in several hours or a ~30Gb hexaploid redwood genome in a few days on a single machine. For trio binning assembly:
| Dataset | Cov. | CPU time | Elapsed time | RAM | N50 |
|---|---|---|---|---|---|
| HG00733, [father], [mother] | ×33 | 269.1h | 6.9h | 135G | 35.1Mb (paternal), 34.9Mb (maternal) |
| HG002, [father], [mother] | ×36 | 305.4h | 7.7h | 137G | 41.0Mb (paternal), 40.8Mb (maternal) |
| NA12878, [father], [mother] | ×30 | 180.8h | 4.9h | 123G | 27.7Mb (paternal), 27.0Mb (maternal) |
Except NA12878, the assemblies above were produced by hifiasm v0.12 and can be downloaded at
ftp://ftp.dfci.harvard.edu/pub/hli/hifiasm/submission/hifiasm-0.12/
NA12878 was assembled with an older version of hifiasm and is available at
ftp://ftp.dfci.harvard.edu/pub/hli/hifiasm/NA12878-r253/
# 帮助
For detailed description of options, please see man ./hifiasm.1. The -h
option of hifiasm also provides brief description of options. If you have
further questions, please raise an issue at the issue
page.
